Paper Publication (Co-authored at Eli Lilly)
mAbs Journal, Volume 14, Issue 1, 2022
mAbs Journal, Volume 14, Issue 1, 2022
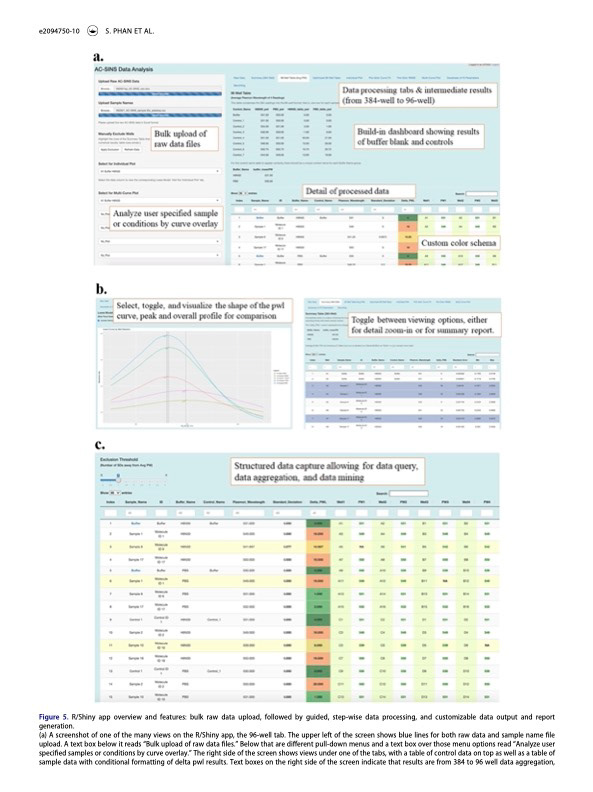
While working as a data scientist at Eli Lilly & Co, I had the wonderful opportunity to develop an R Shiny web application to help automate analysis for an experimental method called AC-SINS (Affinity Capture Self-Interaction Nanoparticle Spectroscopy).
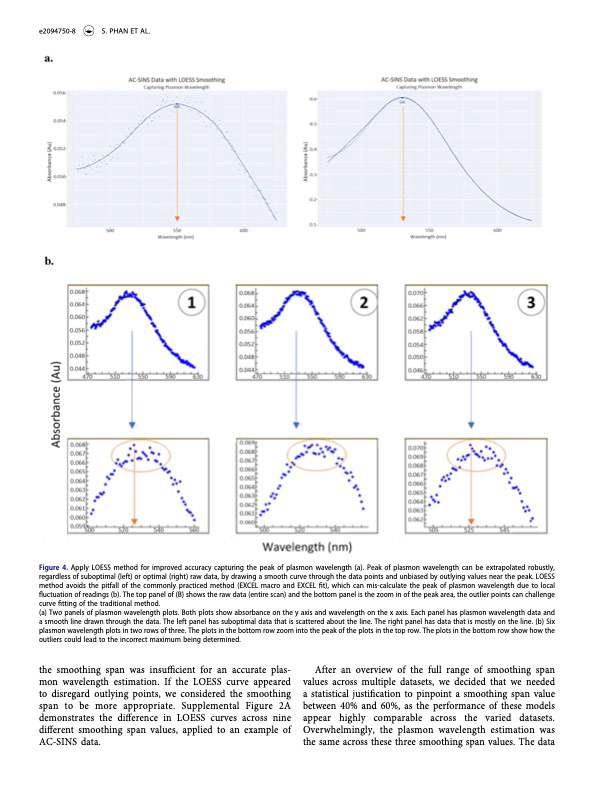
The AC-SINS method allows for early stage characteristic identification of therapeutic antibody candidates. (See my post here about the AC-SINS Analysis Application that I created for Eli Lilly.) Within the process of creating an analysis tool for AC-SINS, my supervisor Eudean Shaw and I arrived at a new analytical approach to automate the extraction of AC-SINS results, using a method called LOESS (local estimated scatterplot smoothing). We worked closely with scientists Qing Chai and Samantha Phan to provide a tool that would serve their analysis purposes and improve the process of conducting the AC-SINS method.
The AC-SINS method allows for early stage characteristic identification of therapeutic antibody candidates. (See my post here about the AC-SINS Analysis Application that I created for Eli Lilly.) Within the process of creating an analysis tool for AC-SINS, my supervisor Eudean Shaw and I arrived at a new analytical approach to automate the extraction of AC-SINS results, using a method called LOESS (local estimated scatterplot smoothing). We worked closely with scientists Qing Chai and Samantha Phan to provide a tool that would serve their analysis purposes and improve the process of conducting the AC-SINS method.
Amidst this project, it was proposed that we write a scientific paper, delving into not only Qing and Samantha’s unique adaptations of the AC-SINS method, but also into the statistical method employed within the analysis application and the unique features of the application itself. We began drafting, exploring why the former method was excessively tedious and inefficient, and why our method was optimal and improved. After many months of collaboration, writing, and creating visualizations to accompany the text, we arrived at a document that we were ready to submit for editing.
I’m very excited to share that as of July 2022, the paper has been published!
The paper is called “High-throughput profiling of antibody self-association in multiple formulation conditions by PEG stabilized self-interaction nanoparticle spectroscopy,” published in mAbs Journal, Volume 14, Issue 1.
You can access it here!
As linked in my AC-SINS Analysis Application post, the GitHub repository with code, documentation, and example data can be found here.
I feel so grateful to my former supervisor, Eudean Shaw, and to Qing Chai and Samantha Phan, who all put a significant amount of trust in me to work with them on this project. I feel honored to be a co-author of a published scientific paper!
The paper is called “High-throughput profiling of antibody self-association in multiple formulation conditions by PEG stabilized self-interaction nanoparticle spectroscopy,” published in mAbs Journal, Volume 14, Issue 1.
You can access it here!
As linked in my AC-SINS Analysis Application post, the GitHub repository with code, documentation, and example data can be found here.
I feel so grateful to my former supervisor, Eudean Shaw, and to Qing Chai and Samantha Phan, who all put a significant amount of trust in me to work with them on this project. I feel honored to be a co-author of a published scientific paper!
Some Example Pages...👇